


In glycobiology and Glycoproteomics Research, accurately deciphering the microheterogeneity of Glycosylation Modifications, i.e., identifying the specific glycopeptides (peptide segment + glycan chain) attached to glycoproteins, is crucial for understanding their function. However, due to the complex structure of glycan chains, their propensity to form adducts, and their unique mass spectrometry fragmentation behavior, glycopeptide data analysis has long been a recognized technical bottleneck.
Although several glycopeptide analysis software programs have emerged in recent years, they still differ in their identification depth, reliability, and ability to handle complex situations. Recently, Protein Prospector, a long-standing software with a 30-year history, has demonstrated significant advantages in glycopeptide identification through a series of innovative improvements.
A study published in Molecular & Cellular Proteomics in February 2025, titled "Improving the Depth and Reliability of Glycopeptide Identification Using Protein Prospector," showed that Protein Prospector can identify a significantly greater number of glycopeptide spectra and glycan types from the same dataset than other software, and can correct a large number of misassignments by considering glycan adducts. This research provides us with a powerful new tool and important insights for selecting and analyzing glycopeptide data.
Glycosylation is the most common and diverse post-translational modification of proteins. In mammals, N-glycosylation is easier to analyze than O-glycosylation due to its defined sequence motif, conserved core pentasaccharide structure, and relatively stable glycosidic bond, but its complexity should not be underestimated.
The main challenges include:
The core breakthrough of Protein Prospector lies in its optimized glycopeptide analysis process, which is not a simple database search, but a layered, mutually verifying three-step strategy:
Using its MS-Filter module, it first filters out all mass spectra containing characteristic fragment ions of N-acetylhexosamine (m/z 204.087), greatly reducing the search scope.
Then, using the main search engine Batch-Tag, a peptide-centric database search is performed in a database containing 730 mammalian N-glycans. This stage primarily relies on peptide backbone fragment ions to identify peptide sequences, and glycan assignment is based solely on mass shift.
The results from the first step are rigorously filtered in Search Compare with a 1% false discovery rate to obtain a high-confidence list of core glycopeptides (peptide + glycan type).
This is the most crucial innovative step. The software uses the core peptide list obtained in the previous step to run MS-Filter again, searching for the corresponding Y1 ions of these peptides in the entire dataset.
The Y1 ion is a peptide ion that retains a core GlcNAc after glycopeptide fragmentation, and its mass is equal to "peptide mass + 203.08 Da". Finding the Y1 ion means finding another possible glycopeptide variant of that peptide carrying a different glycan chain.
For each candidate spectrum found, the software not only confirms the Y1 ion but also systematically scores the glycan fragment ions, evaluating the match between the observed B/Y ions and the hypothetical glycan structure, thus selecting the most likely one from multiple candidate glycan types.
This strategy of first locking onto the peptide and then using it as an anchor to search for more glycan types cleverly bypasses the difficulty of directly identifying all complex spectra from scratch, greatly improving identification sensitivity and efficiency.
To objectively evaluate performance, researchers used a publicly available Mass Spectrometry dataset of mouse liver glycopeptides and conducted a comparative analysis using six mainstream software programs (Byonic, GlycoDecipher, MSFragger, pGlyco3, Protein Prospector, StrucGP).
The results were impressive:
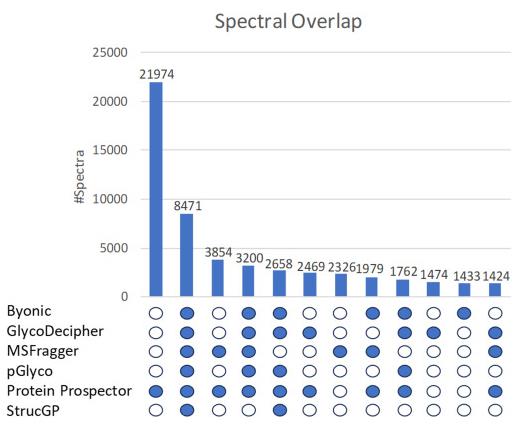
Fig. 1 Overlap of spectral identification results from six glycopeptide analysis software programs on a mouse liver dataset. (Chalkley, et al. 2025)
Adducts, particularly ammonium adducts, are an easily overlooked yet significantly impactful factor in glycopeptide analysis. This study reveals the dual role of considering adducts:
The study also highlights common challenges in current analyses.
Background glycan fragment ions are ubiquitous. Even in glycopeptide spectra identified as not containing NeuGc (N-glycolylneuraminic acid), over 70% showed characteristic NeuGc fragment ions (m/z 290, 308). This is not due to the presence of NeuGc in the glycan, but rather from co-isolation or background interference. This requires analytical software with strong interference-resistant scoring capabilities.
Peptide-first and glycan-first strategies each have their advantages.
This study confirms that Protein Prospector, through its unique three-step analysis process, particularly the glycan extension search based on Y1 ions, can extract deeper and broader glycopeptide information from complex data.
The implications for glycoproteomics researchers are:
In the future, as the software further integrates glycan topological structure inference functions and continuously optimizes algorithms for handling background noise and adducts, we expect to extract more accurate and complete glycosylation information from valuable mass spectrometry data, thereby revealing the mysteries of glycobiology in health and disease in greater depth.
Reference








