The precisION Platform Enables In-Depth Analysis of Protein Modifications
January 14, 2026
Post-translational modifications of proteins (such as phosphorylation, Glycosylation, and lipidation) are central to their functional regulation. However, accurately capturing these modifications in intact protein complexes, especially when they are masked by numerous glycan chains, has been a technical challenge.
On September 29, 2025, a team from the University of Oxford, led by Carol V. Robinson and Corinne A. Lutomski, published their research findings in Nature, titled "Uncovering hidden protein modifications with native top-down mass spectrometry." The researchers developed an interactive open-source software called precisION, which combines an innovative fragment-level open search algorithm to perform in-depth top-down analysis of intact protein complexes under non-denaturing mass spectrometry conditions. This allows for the discovery, localization, and quantification of previously hidden chemical modifications with unprecedented precision.
Why are modifications in intact complexes so difficult to detect?
Traditional bottom-up proteomics requires enzymatic digestion of proteins into peptides. While this can identify modifications, it destroys the protein's intact structure and complex information, preventing direct correlation of specific modifications with the protein's higher-order structure and interactions.
Non-denaturing top-down mass spectrometry (nTDMS) technology can directly analyze intact proteins and their complexes, performing fragmentation analysis while maintaining their native state. Theoretically, this perfectly solves the above problems. However, in practice, a large amount of crucial information remains hidden due to the following reasons:
- High heterogeneity: Glycoproteins often carry heterogeneous modifications, resulting in broad and complex mass spectrometry peaks, making it impossible to resolve individual modifications at the intact protein level (MS1).
- Low fragmentation efficiency: Under non-denaturing conditions, proteins have lower charges, and fragmentation is less efficient than under denaturing conditions, resulting in weaker fragment ion signals.
- Database dependence: Conventional analytical methods rely on databases of known modifications, making it difficult to discover uncharacterized or novel modifications.
The Core Technological Breakthrough of precisION
The precisION software package builds a powerful bioinformatics framework, whose core innovation lies in its fragment-level open search algorithm. It no longer relies on known intact protein masses or modification databases, but directly analyzes all fragment ions produced after fragmentation. Its workflow is like a precise molecular puzzle:
- High-quality deconvolution: Accurately extracts isotope peak clusters of fragment ions from complex raw mass spectra.
- Protein identification: Determines the identity of the analyzed complex through de novo sequencing or open database searching.
- Hierarchical fragment matching: Systematically matches all unmodified fragment ions first.
- Open search for modification discovery: For the remaining large number of unmatched fragments, an open search algorithm is applied to find sets of fragment ions with common mass offsets. This offset represents a modification (e.g., a phosphate group +80 Da, or a glycosyl group + a specific mass). The algorithm statistically evaluates the significance of these offsets, thus discovering and locating novel modifications.
This method can not only discover small modifications (such. as phosphorylation) but also identify large modifications through multi-gap searching, such as protein truncation or internal fragments with large missing segments, enabling comprehensive and in-depth mapping of Protein Modifications.
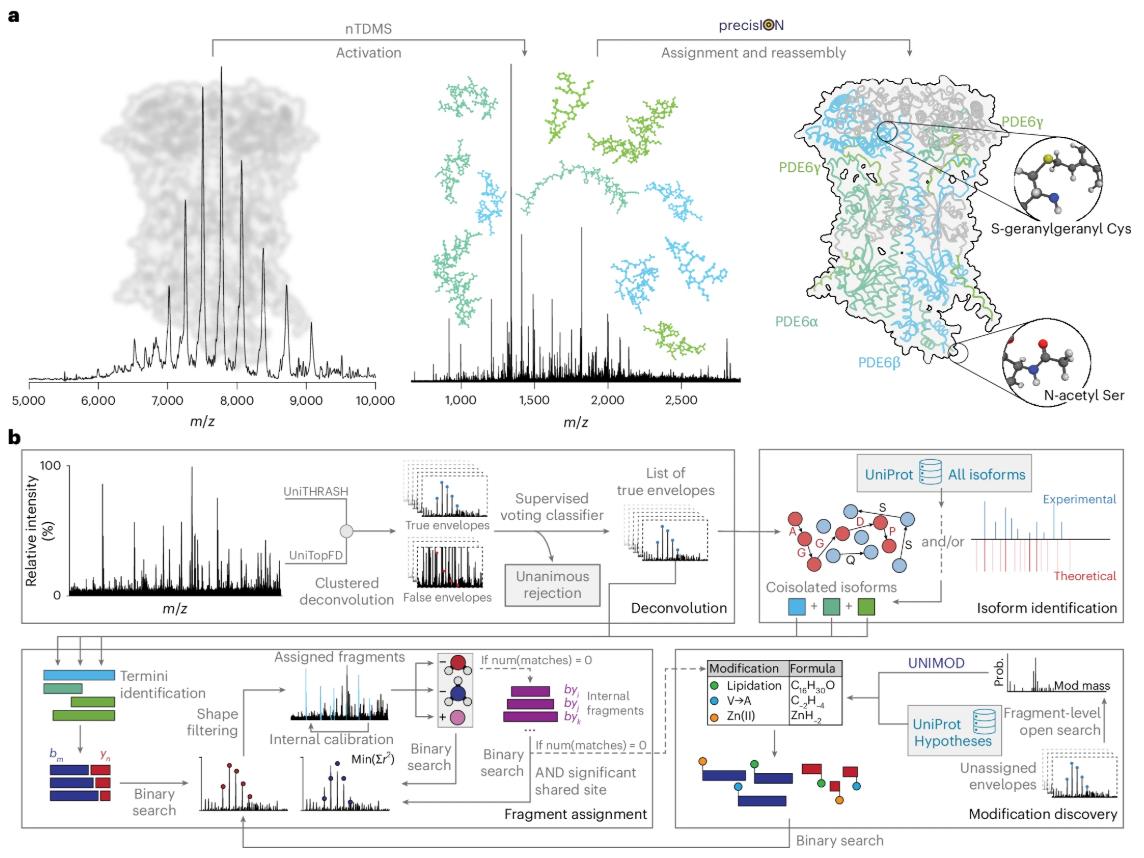
Fig. 1 Data-driven interpretation of native top-down mass spectra in precisION. (Bennett, et al. 2025)
Unveiling Underlying Biology
The research team applied precisION to several protein complexes with significant biological and clinical importance, achieving a series of groundbreaking discoveries.
Precise Localization of N-glycosylation on the ACE2 Receptor
Human angiotensin-converting enzyme 2 (ACE2) is a key receptor for the novel coronavirus, and its surface has six potential N-glycosylation sites. The glycosylation state affects its dimerization and function. Even using cell lines expressing homogeneous glycoforms, the mass spectra of ACE2 dimers remain complex.
- precisION analysis of its fragment spectra not only confirmed the occupancy of the main glycosylation sites but also provided the first direct observation of variable proteolytic processing at the C-terminus of ACE2.
- Through statistical analysis of internal fragment sets, the sequence coverage was significantly increased from 10.6% to 30.9%.
- The complete glycan chain (Hex5HexNAc2) and partially degraded Glycan Chains (such as a single HexNAc) were directly detected at the fragment level, still attached to specific peptide fragments, providing new insights into the fragmentation behavior of glycoproteins in the gas phase.
Unveiling the Modification Hierarchy of the Signaling Glycoprotein SPP1
Osteopontin (SPP1) is an extracellular matrix protein that is heavily O-glycosylated and phosphorylated, exhibiting high modification heterogeneity and diverse functions.
- Against a broad background of O-glycosylation, precisION successfully penetrated the interference of glycosylation.
- Two previously unknown C-terminal truncated variants of SPP1 were discovered, and their possible protease sources were inferred.
- Although SPP1 has more than 50 potential phosphorylation sites, precisION's quantitative analysis showed that the phosphorylation level in this recombinant protein was generally low and mainly enriched in specific regions. This finding is consistent with the overall phosphorylation level after deglycosylation.
Deciphering the Diverse Lipid Modifications of the Neurotransmitter Transporter GAT1
GABA transporter 1 (GAT1) is responsible for the reuptake of the inhibitory neurotransmitter GABA and is an important target for anti-epileptic drugs.
- Intact protein mass spectrometry showed mass increases of approximately +238 Da and +265.5 Da in GAT1, suspected to be due to lipid modification, but difficult to confirm.
- precisION's fragment-level open search clearly identified these shifts as corresponding to various fatty acid modifications, including palmitoylation (16:0), oleoylation (18:1), and stearoylation (18:0), and quantified their relative abundance.
- By comparing the positions of modified and unmodified fragments, the lipid modification was precisely localized to Cys493 (rather than the nearby Cys499).
- Most remarkably, the researchers re-examined the cryo-EM structure of GAT1 and found an unassigned electron density near Cys493. Fitting the palmitic acid chain model identified by precisION into this density resulted in a perfect match, revealing its interaction with a phospholipid molecule (phosphatidylethanolamine), providing direct structural evidence for understanding the role of lipid modification in Membrane Protein Localization and Function.
Opening a New Chapter in Integrative Structural Biology
The development of precisION marks a new data-driven, discovery-oriented stage in nTDMS data analysis. Its significance lies in:
- Technological level: It provides a powerful, open-source, and user-friendly platform that transforms complex fragmentation mass spectra into interpretable biological insights, greatly unleashing the potential of nTDMS technology.
- Glycobiology level: It provides unprecedented tools for studying the microheterogeneity of glycosylation at the intact complex level, the crosstalk between glycosylation and other modifications (such as phosphorylation), and the direct impact of glycosylation on protein structure and interactions.
- Structural Biology Level: It enables deep integration of mass spectrometry with techniques such as cryo-electron microscopy. Covalent modifications discovered by mass spectrometry (such as the lipidation of GAT1) can provide crucial clues for interpreting electron microscopy density maps, thereby constructing more accurate atomic models closer to the physiological state.
In the future, precisION is expected to drive the shift from depicting static structures to dynamically analyzing functional protein modification networks. This will not only deepen our understanding of fundamental biological processes but also provide new avenues for discovering disease-related specific modified protein forms (proteoforms) and developing more precise drugs.
Related Services
Reference
- Bennett, J. L., et al. (2025). Uncovering hidden protein modifications with native top-down mass spectrometry. Nature Methods, 1-11. DOI: 1038/s41592-025-02846-5.












