


On February 20, 2025, a team led by Richard D. Cummings from Harvard Medical School published a study in the Journal of Proteome Research titled "Swift Universal Glycan Acquisition (SUGA) Enables Quantitative Glycan Profiling across Diverse Sample Types," introducing a novel Glycomics Analysis method called Swift Universal Glycan Acquisition (SUGA), which enables rapid, high-throughput, and label-free glycan analysis and quantification.
Glycans are complex biomolecules composed of Monosaccharides linked by glycosidic bonds. They combine with proteins or lipids to form glycoproteins or glycolipids. Glycosylation plays a crucial role in almost all life processes, including cell recognition, signal transduction, and immune response. Abnormal Glycosylation is closely associated with cancer, autoimmune diseases, and infectious diseases.
Therefore, comprehensively and rapidly analyzing the glycan composition in biological samples is crucial for understanding disease mechanisms and developing new biomarkers or drugs.
Current mainstream Glycan Mass Spectrometry Analysis workflows typically include: enzymatic release of glycans from glycoproteins, reduction (opening the reducing end cyclic structure), derivatization (such as methylation to improve ionization efficiency or attachment of fluorescent labels), liquid chromatography separation, and finally mass spectrometry detection.
These steps are not only time-consuming (up to several hours per sample), but the complex chemical processing may introduce bias or lead to the loss of some fragile glycans (such as sialic acids). Throughput bottlenecks severely limit the application of glycomics in large-scale clinical studies or high-throughput drug screening.
SUGA is a rapid glycan analysis method based on Direct Injection-Electrospray Ionization Mass Spectrometry (Direct Injection-ESI-MS), with the following characteristics:
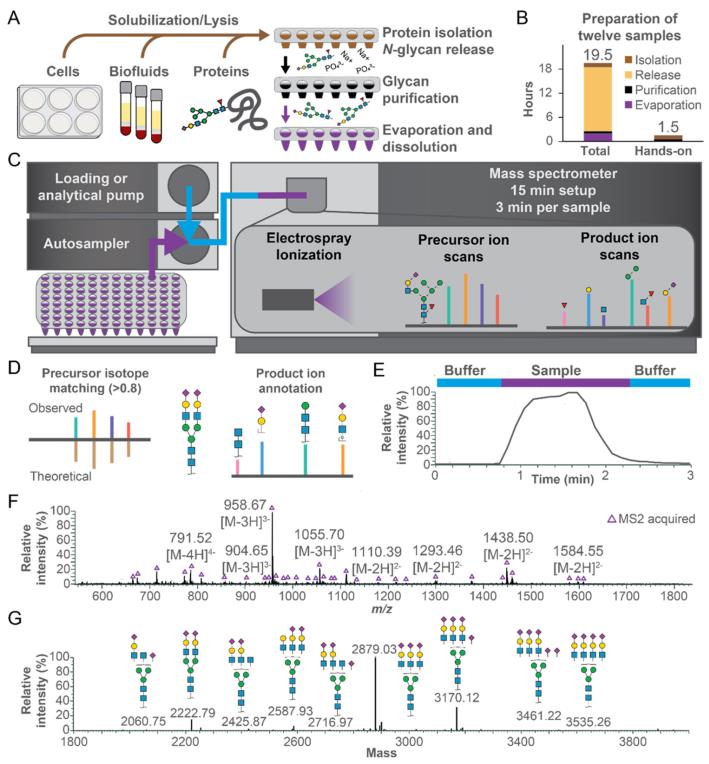
Fig. 1 Overview of Swift Universal Glycan Acquisition (SUGA) by Data Dependent Acquisition Mass Spectrometry (DDA-MS) method. (A) Sample preparation workflow was used to obtain N-glycans for SUGA. (B) Duration of the sample preparation steps. (C) Instrument setup for automated sample injection and data acquisition. (D) Criteria used for high-confidence glycan identification. Native bovine fetuin N-glycans were analyzed by SUGA. (E) Total Ion Chromatogram (TIC). (F) Negative mode precursor ion mass spectrum with monoisotopic masses and ions subjected to fragmentation labeled. (G) Charge deconvoluted precursor ion mass spectrum with the corresponding glycan identifications. (Ashwood, et al. 2025)
To verify the reliability of SUGA, the research team systematically compared it with the currently widely accepted gold standard for glycan analysis (MALDI-TOF MS Analysis after methylation derivatization).
The researchers selected five purified glycoproteins with different types of N-glycans (high-mannose type, complex neutral type, and complex sialylated type). The same released glycan sample was divided into two portions: one was analyzed using SUGA for natural glycans, and the other was methylated and then analyzed using both SUGA and MALDI-TOF.
The results showed:
For direct injection, a separation-free method, concerns naturally arise regarding its technical reproducibility. Researchers evaluated the variability of SUGA by performing five replicate experiments using ExpiCHO cell lysate.
Within a dynamic range of 2.5 orders of magnitude, only 2 of the 58 detected glycan compositions had a CV greater than 20%, with a median CV of approximately 10%. The coefficient of variation was negatively correlated with the signal intensity of the glycans, meaning that stronger signals resulted in better reproducibility.
Simultaneously, the study investigated the impact of the initial protein amount on the results. It was found that when the initial protein amount was below 50 μg, not only did the glycan signal intensity decrease, but the CV of some low-abundance glycans increased, and even the abundance ranking of glycan compositions began to be distorted. Therefore, it is recommended to use at least 50 μg of initial protein for glycan release to ensure data stability.
To demonstrate SUGA's powerful capabilities in high-throughput, quantitative analysis, researchers applied it to unbiased screening of Glycosylation Inhibitors.
They treated ExpiCHO cells with three inhibitors with different mechanisms of action:
These experiments demonstrate that SUGA can unbiasedly capture changes in glycan composition and reveal the temporal dynamics of different inhibitor actions, providing a powerful tool for glycosylation pathway research.
Of course, the SUGA method currently has its limitations:
Nevertheless, SUGA has broad prospects. It provides an elegant and powerful solution to address the high-throughput bottleneck in glycomics. With further optimization and popularization of the method, it is expected to:
The emergence of the SUGA method represents a significant innovation in the field of glycomics analysis. It reduces the analysis time for a single sample from hours to minutes, achieving a leap in throughput while maintaining data quality.
Through direct analysis without derivatization, SUGA not only simplifies the process and increases speed but also reduces potential interference with fragile glycan structures. Its high consistency with the gold standard method and its powerful quantitative capabilities demonstrated in high-throughput inhibitor screening fully prove its reliability, robustness, and practicality.
As the role of glycobiology in life and health and disease treatment becomes increasingly prominent, efficient and agile analytical tools like SUGA will undoubtedly become a key tool for researchers to unlock the mysteries of glycobiology, driving the entire field to develop rapidly in greater depth and breadth.
Reference








